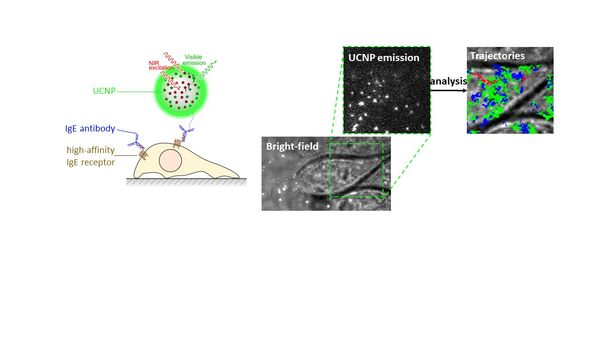
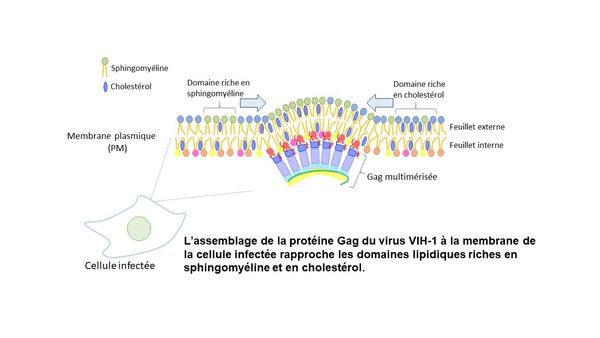
Le Laboratoire de Bioimagerie et Pathologies
L’objectif du Laboratoire de Bioimagerie et Pathologies est, par une approche multidisciplinaire (biologie/chimie/physique) et multi-échelle (de la molécule au patient), d’étudier à un niveau fondamental et appliqué les propriétés et les rôles d'un nombre limité de biomolécules clés impliquées dans différents processus pathologiques. Notre approche consiste à développer, sur la base d’informations acquises à un niveau fondamental, de nouvelles approches thérapeutiques et diagnostiques en cancérologie et en microbiologie. Une particularité clé de l’unité est de développer et caractériser en parallèle des outils, méthodes et techniques innovants basés sur la fluorescence pour répondre à ces questions. Cet ensemble d’approches pluridisciplinaires qui associe des développements d’outils fluorescents et d’imagerie innovants au service de problématiques biologiques constitue une singularité dans le paysage scientifique français.